
Pierre Baduel
Polyploidy, Epigenetics & Transposable Elements @IBENS

- Research -
My research focuses on understanding the contribution of transposable elements (TEs) as a source of heritable allelic and epiallelic variation of functional importance and how it can be modulated by genetic as well as environmental determinants (theme 1), mating systems (theme 2), or ploidy (theme 3). To tackle these questions, I use the species of the Arabidopsis genus, notably the model plant A. thaliana, as a study system and combines multi-omic bioinformatic approaches with plant molecular genetics and field collections.
Theme 1 - Genetic and environmental determinants of transposition in nature
Standing genetic variation is generally thought to be the main source of rapid adaptation to environmental changes. As a result, genomic studies aimed at estimating the evolutionary potential of native populations in future climates have mostly focused on SNPs. However, there is increasing evidence that the rare and typically large effect alleles created by TE insertions, even though mostly deleterious, can be sometimes beneficial and contribute significantly to local adaption. Leveraging the multi-omic and bio-climatic data for more than 1,000 wild A. thaliana accessions sequenced as part of the 1001 Genomes project, we set out to determine the rate of TE mobilization and its potential to create adaptive variation in natural settings.
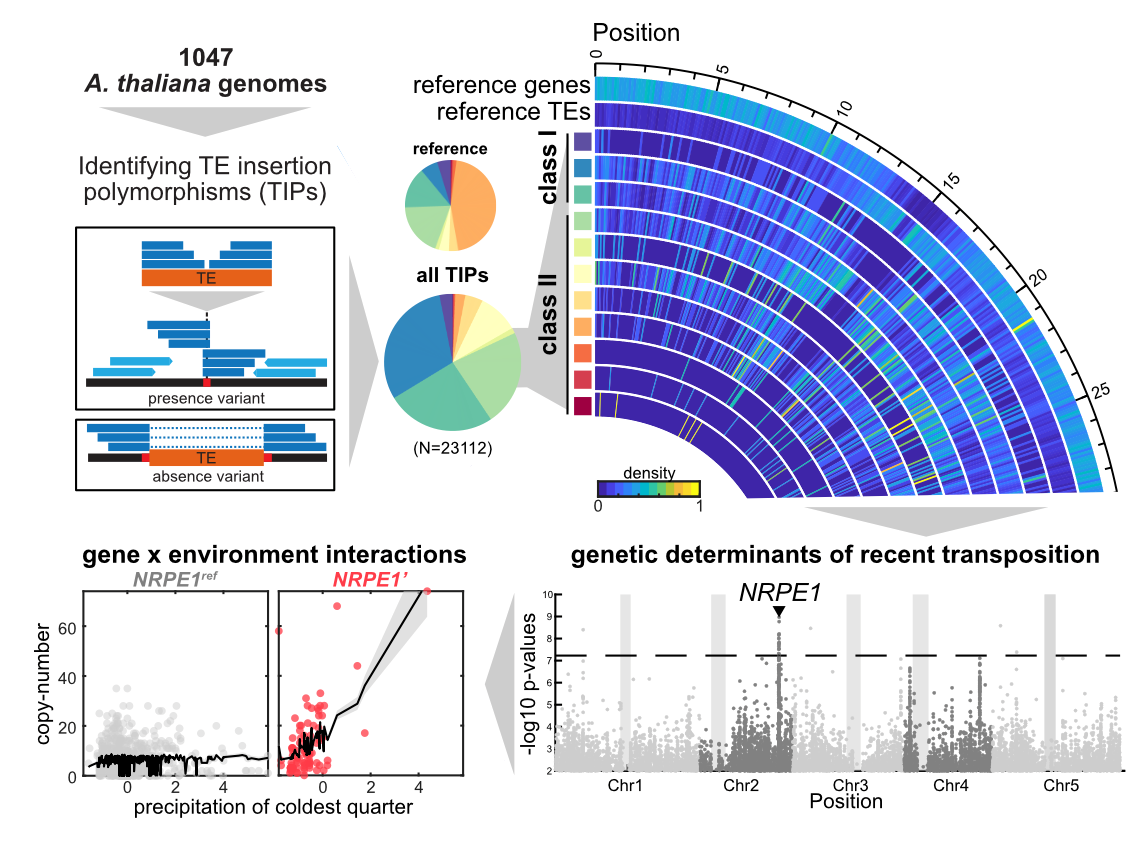
Using an updated pipeline to detect TE insertion polymorphisms (TIPs) from short-read sequencing data (see SPLITREADER package here), we identified >23,000 TIPs across >1000 accessions from across the globe. Based on this dataset, we determined the substitution rate of TIPs in nature to be almost a third of that of SNPs (between 0.06-0.08 TE insertion per genome per generation) and very close to the actual transposition rate we measured experimentally. However, unlike SNPs, TE insertions tend to have large deleterious effects when they insert within or near genes (which occurs at least 50% of the time for some TE families) and are therefore rapidly purged by purifying selection. Nonetheless, we found some gene loci to be recurrently targeted by TE insertions (most notably FLC’s 1st intron), and these tended to be found in disturbed environments where they showed signatures of positive selection.
Furthermore, we also found that a naturally truncated variant of NRPE1 (a key component of the RdDM pathway which targets DNA methylation over TEs) causes lower CHH methylation and higher levels of transposition, largely by enhancing the environmental sensitivity (GxE) of TEs. Strikingly, this mutator NRPE1 allele is enriched at the edge of the environmental niche of A. thaliana where it itself shows signature of positive selection, akin in a way to bacterial mutator alleles that can be temporarily beneficial in harsh environments. How such an allele has been retained over long term in A. thaliana opens exciting avenues of inquiry, especially in times of climate crisis where TEs may be essential genomic players in the demise or rescue of native populations.
Related publications
Baduel P, Leduque B, Ignace A, Gy I, Gil J, Loudet O, Colot V, Quadrana L. Genetic and environmental modulation of transposition shapes the evolutionary potential of Arabidopsis thaliana. Genome Biology, 10.1186/s13059-021-02348-5, 05/2021.
Baduel P, Quadrana L. Jumpstarting evolution: How transposition can facilitate adaptation to rapid environmental changes. Current Opinion in Plant Biology, 10.1016/j.pbi.2021.102043, 04/2021.
Baduel P and Sasaki E. The genetic basis of epigenetic variation and its consequences for adaptation. Current Opinion in Plant Biology, 10.1016/j.pbi.2023.102409, 08/2023.
Theme 2 - Impact of the mating type on TE mobilization dynamics
Theoretical considerations predict that TE dynamics are shaped in large part by the mating system (Wright and Schoen 1999), which in plants switches recurrently from outcrossing to selfing (Shimizu and Tsuchimatsu 2015). However, the direction of this effect remains unclear as changing mating system can have a number of consequences with potentially antagonizing influences on the dynamics of TEs (Boutin et al. 2012). On the one hand, selfing increases homozygosity compared to outcrossing, and thus exposes to purifying selection recessive deleterious alleles, such as those caused by most TE insertions (Arunkumar et al. 2015). On the other hand, the effective population size of selfers is expected to be drastically lower as compared to that of outcrossers, hence decreasing the efficacy of purifying selection against deleterious mutations (Pollack 1987). In addition, selfing is expected to limit the invasion of active TEs between genomes while outcrossing may favor instead an epidemic-like spread (Cavalier-Smith 1980). Finally, although the increased level of homozygosity in selfers results in lower effective homologous recombination rate, the actual frequency of crossovers is expected to be higher in selfers than in outcrossers (Roze and Lenormand 2005). How these different effects impact TE accumulation and ectopic recombination between dispersed TE copies is not known. Therefore, while theory predicts that mating systems play important roles in shaping TE dynamics (Wright et al. 2008), the net outcome of the balance between different forces, i.e. the nature of the deleterious effects of TEs (through recombination based processes or direct deleterious disruption of gene function) and their dominance/recessivity is generally unresolved.
To study how mating systems impact the dynamics of TE accumulation, we used the outcrossing species Arabidopsis lyrata which also has some North American populations that have recently experienced a shift to selfing. Specifically, we have sequenced from both selfing and outcrossing populations 20 genomes using long reads Oxford Nanopore Technology as well as 19 additional ones using short read Illumina sequencing. Using the short reads sequencing data, we have characterized the impact of selfing on genetic diversity as well as on the efficacy of purifying selection at the Single Nucleotide Polymorphism (SNP) level. After assembling the long-read sequenced genomes de novo, we have then detected transposable element insertion polymorphisms (TIPs) across these populations.
Funding This axis of research is part of the ANR PRC TE-MoMA (AAPG PRC 2018) led by Vincent Castric (Lille University) in collaboration with Vincent Colot (IBENS) and Jean-Marc Aury (Genoscope).
Theme 3 - Impact of autopolyploidy on the mobilization dynamics of TEs
Whole genome duplications (WGDs) are recurrent events throughout eukaryote evolution and the initial adaptive advantage they are associated with has been attributed in part to the increased genetic diversity of polyploid genomes (Baduel et al., Front Ecol Evol 2018). Yet, the molecular mechanisms generating this diversity remain unclear.
During my PhD in the group of Kirsten Bomblies at Harvard University, I set out to determine the impact of WGD on adaptation and evolution using Arabidopsis arenosa as a model. This species is a close relative of A. thaliana that exists in two ploidy forms: an ancestral diploid form and a derived autotetraploid form resulting from a WGD event that took place ~60,000 years ago. As the two forms are usually sympatric, A. arenosa tetraploids and their diploid progenitors provide an ideal system to infer, without the confounding effect of interspecific hybridization, the consequences of polyploidization per se on adaptation.
Focusing on a tetraploid population of A. arenosa that has successfully colonized the railway tracks of Central Europe, a habitat highly disturbed by humans, I identified the molecular basis of two key traits associated with this invasion, a constitutive stress tolerance (Baduel et al., Plant Phys 2016) and a switch to rapid-cycling and early-flowering (Baduel et al., PLoS Gen 2018). Following these studies and taking advantage of a large genome sequencing project of ~300 A. arenosa individuals across both ploidies, which provided the first genome-wide validation of the increased nucleotidic diversity of tetraploid genomes, I then showed that this increased genetic diversity is due to the combined effect of an increased effective population size (double the number of genome copies per individual) and the relaxed purifying selection associated with polysomic masking (Monnahan, Kolar, Baduel* et al., Nat Ecol Evol 2019).
In contrast to small nucleotidic variants which are mostly neutral or weakly deleterious, the mutations that transposable elements (TEs) generate when they insert near or within genes tend to have large-effect, most of which are deleterious and under strong purifying selection. I therefore predicted that the relaxed selection I measured in A. arenosa tetraploids may significantly impact the accumulation dynamics of TEs.
Using a bioinformatic pipeline to detect TE insertions from short-read sequencing data developed in the Colot group and that I subsequently improved (Baduel et al., Meth Mol Biol 2021), I was able to compare TE accumulation in diploid and tetraploid A. arenosa genomes. My results unequivocally indicate that, consistent with my prediction, the relaxation of purifying selection in tetraploids is a major factor of increased TE load. Even though it has long been hypothesized that WGD represented a genomic shock that would trigger a transposition burst, I did not detect any such burst, suggesting that this type of response, if it exists, is not systematic and perhaps requires specific genetic or environmental factors.
Furthermore, I found that the TE insertions that accumulated within polyploid genomes, notably near or within genes involved in environmental response, could prove beneficial in new environments. This was notably the case for railway tetraploids which carried a TE insertion disrupting the coding sequence of FLOWERING LOCUS C (FLC), the major flowering repressor gene that I had identified as one of the key loci underlying the switch to early-flowering. These results indicate that, at least in the short-term, over-accumulation of TEs can contribute to the rapid adaptation of polyploids to novel environments (Baduel et al., Nat Comm 2019). Yet, as TE sequences represent a major substrate for ectopic recombination, I argued that their accumulation could also contribute to the long term evolutionary decay of polyploid genomes by promoting their re-diploidization (Baduel et al., Front Ecol Evol 2018).
Related publications
Baduel P, Quadrana L, Hunter B, Bomblies K, Colot V. Relaxed purifying selection in autopolyploids drives transposable element over-accumulation which provides variants for local adaptation. Nature Communications,10.1038/s41467-019-13730-0, 12/2019
Baduel P, Bray S, Vallejo-Marin M, Kolář F, Yant L. The ‘Polyploid Hop’: shifting challenges and opportunities over the evolutionary lifespan of genome duplications. Frontiers in Ecology and Evolution, 0.3389/fevo.2018.00117 07/2018